COM – Le LCB collabore avec le YouTubeur Tanguy Leroux sur la Biodiversité d’un vignoble !
Quels éléments faut-il réunir pour faire un bon vin ? Entre le savoir-faire du vigneron, la qualité des cépages et même la composition des sols
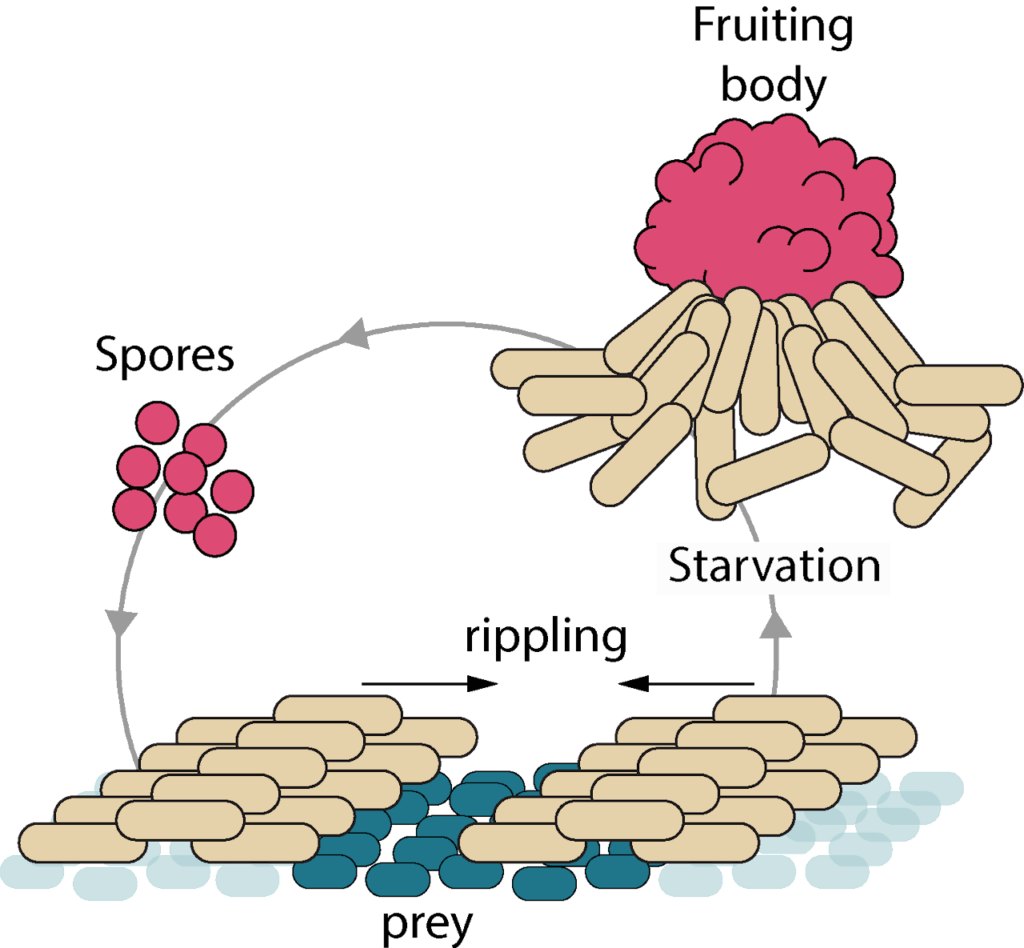
Our team studies the molecular, cellular and evolutionary bases of bacterial predation using Myxococcus xanthus as a model organism. M. xanthus is a ubiquitous soil bacterium that forms coordinated cell groups to invade prey communities by motility and consume them to grow and form multicellular fruiting bodies that host sporulation when the nutrients are exhausted. This is a powerful model system to study how single cell behaviors are regulated spatially and temporally to elicit complex emergent group behaviors, to detect, kill and form multicellular assemblies. To tackle these problems, we take an integrated approach using molecular and single cell systems and linking them to community scales by global computational and theoretical approaches. This research relies on a large collaborative network involving molecular, cellular and structural biologists, biophysicists, mathematicians and bioinformaticians. Specifically, we have developed a unique image analysis framework to observe molecular processes in single cells and map them spatially to the multicellular community.
In the longer term, we hope to bridge these studies to field studies in an attempt to describe how molecular mechanisms defined in the laboratory relate to biological processes that these predators experience in their true ecological niche, where they likely interact with other bacterial species, fungi, nematodes and also perhaps plants.
Beyond the fundamental aspects of this research, there are numerous potential applications, for health (via the discovery of new antimicrobials) but also possibly for agronomy and current environmental issues, at a time where biodiversity is critically endangered by human activity and global warming.
Quels éléments faut-il réunir pour faire un bon vin ? Entre le savoir-faire du vigneron, la qualité des cépages et même la composition des sols
La motilité bactérienne sur surface joue un rôle crucial dans divers processus écologiques et pathogéniques. Dans leur environnement naturel, cette capacité permet aux bactéries de coloniser de
Au terme des élections ouvertes en 2023, l’Académie des sciences vient d’élire 18 nouveaux membres. Parmi ces nouveaux membres, Tâm Mignot, directeur du LCB, a
Tâm Mignot reçoit la médaille d’argent du CNRS. La médaille d’argent distingue des chercheurs et des chercheuses pour l’originalité, la qualité et l’importance de leurs
Découvrez notre deuxième episode des Chroniques Microbiennes ! A l’instar d’autres organismes comme les fourmis ou les abeilles, certaines bactéries peuvent avoir des modes de vie
Dans le cadre de l’année de la biologie (2021-2022), Tâm Mignot, directeur de recherche CNRS au sein du LCB à l’Institut de Microbiologie de la
En cette matinée de Mars 2022, Jade est d’humeur maussade. Vaccinée contre le virus du Sars-Cov-2, elle ne comprend pas comment elle a pu contracter
LCB, in collaboration with idylle have developed Chitozen : The 1st functionalized microscope coverslip to image live bacterial cells & study their growth and behavior

How are Myxococcus cells growing on prey? How do Myxococcus cells uptake nutrients from preys and how is the cell cycle coupled to prey killing and motility? We have designed tools to directly image all the cell cycle stages when Myxococcus is in contact with the prey cells and thus observe cell growth directly at the single cell levelinside the prey fields. This methodology also allows the direct observation of subcellular processes, such as assembly of the cytokinetic ring, chromosome replication and segregation. We also monitor global gene expression changes at various stages of the predation cycle, informing on the metabolic pathways that become activated during predation. We are now developing genetic screens to identify regulatory checkpoints that couple metabolism to the cell cycle upon prey nutrition.
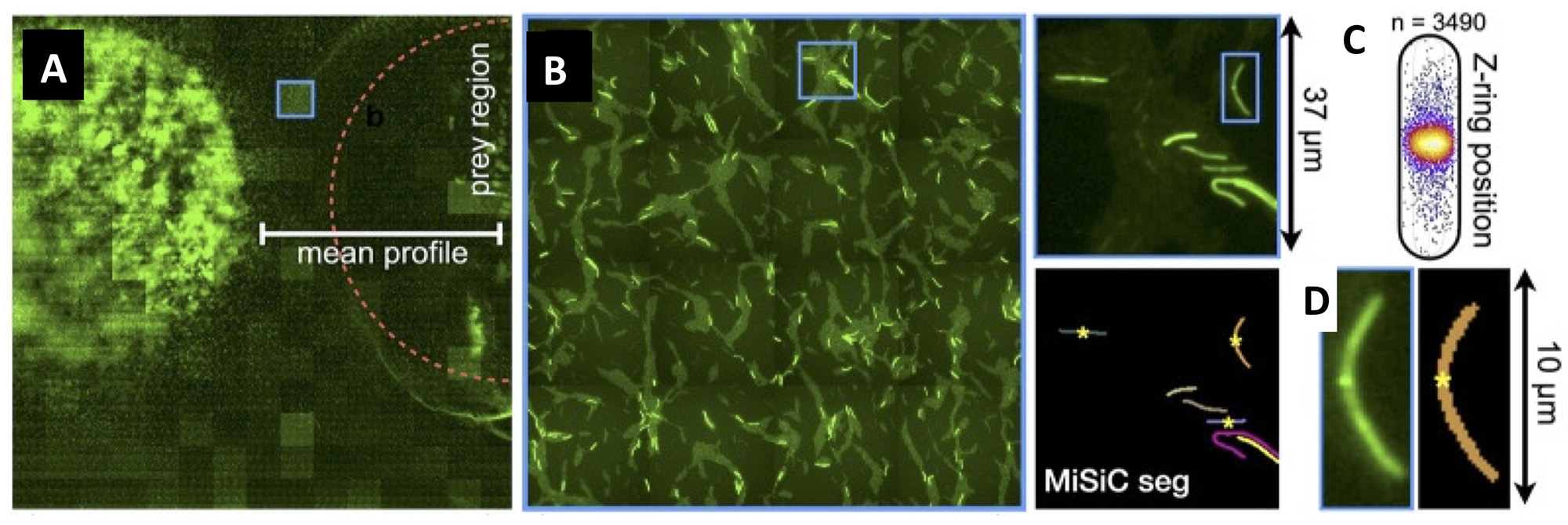
Figure legend : Mapping of M. xanthus cell division in the M. xanthus-E. coli community. (A-B) Bacto-Hubble image of a predatory field containing FtsZ-NG labeled Myxococcus xanthus cells and unlabeled Escherichia coli prey cells. The composite image results from the assembly of 15 × 15 Tile images. The dotted circle marks the limits of the original prey colony. The blue square marks the cell shown as an example in (C). (C) Detection of dividing cells. FtsZ-NG fluorescent clusters are detected at midcell. The FtsZ clusters can be detected as fluorescence intensity maxima. Shown is a projection of the position of fluorescence intensity on a mean cell contour for a subset of n = 3490 cells The blue square marks the cell shown as an example in (C). (D) The example shows segmentation of the field shown in (D). The position of Z-ring foci detected as fluorescent maxima was linked to all fluorescent cells segmented in the MiSiC mask.
How do Myxococcus cells move? How do they make directional decisions and how do coordinated cell groups move and form patterns? Myxococcus cells move in groups (S motility) or in isolation (A motility) by two independent motility complexes: a Type-IV pilus operates Smotility, while the recently discovered Agl-Glt complex drives A motility. We study how both systems act together to promote prey invasion. In particular, both motility systems operate under a common regulatory network, whereby a single regulator, the Ras-like GTPase MglA activates each system at the bacterial cell pole. We study how this protein activates each of the motility systems and how it discriminates them depending on the cellular context.
Last, MglA can be switched to the opposite pole by the activity of a chemosensory-like system (Frz). Consequently, the cells change direction and move in the opposite direction (reversal). We study how activation of the Frz receptor triggers a phosphorylation cascade that switches the Mgl system and we use a combination of high-throughput tracking and modeling to connect these regulations to multicellular behaviors, especially during prey invasion and the formation of rippling waves.
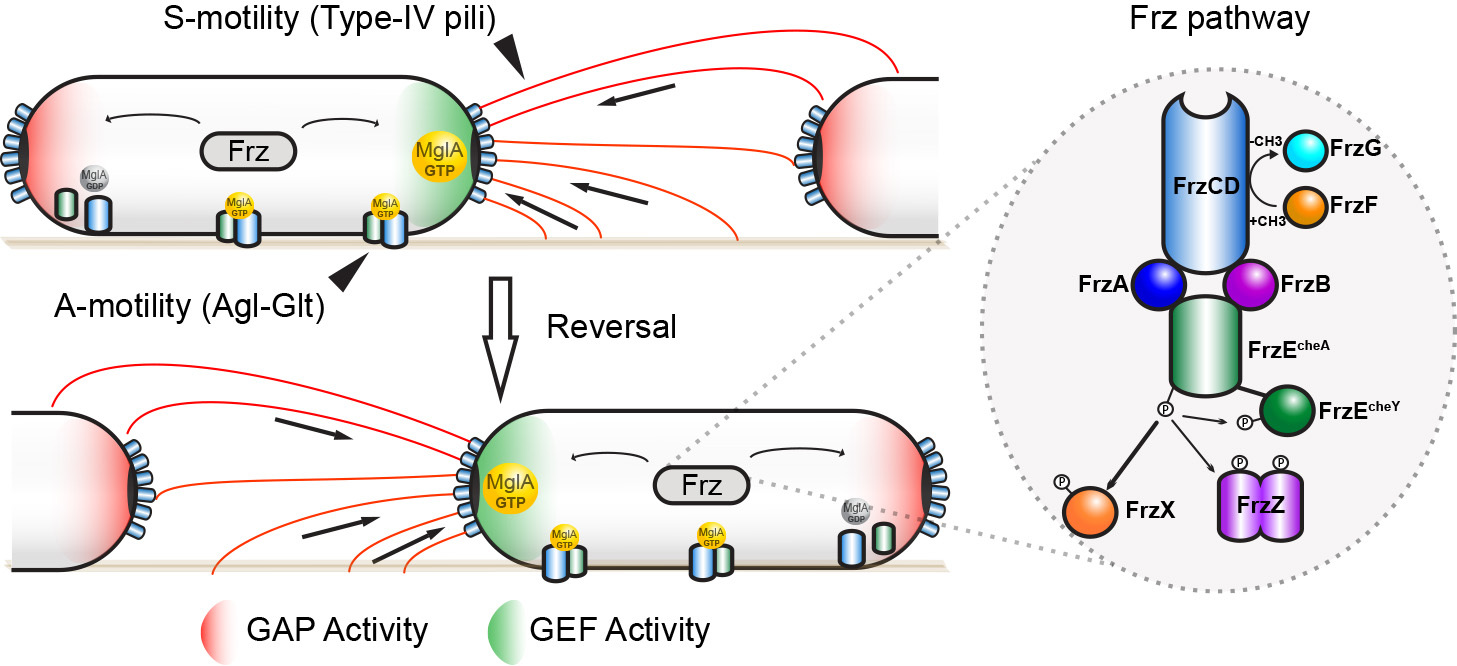
Figure legend : A model for MglA activation by the Frz system in M. xanthus. MglA-GTP is asymmetrically localized at the leading pole of the cell (by the combined action of MglB (GAP Activity, Red) and RomR/X (GEF Activity, Green). Frz proteins are homologues of chemotaxis (Che) proteins. FrzCD is a receptor (MCP), FrzE is a histidine kinase (CheA) coupled to a response regulator domain (CheY), FrzA and FrzB and FrzA are coupling proteins (CheW), FrzX and FrzZ are response regulators (CheY), FrzF is a methyltransferase (CheR) and FrzB a methylesterase (CheB). High Frz activity, in response to yet unknown signals, is translated into high level of phosphorylated FrzX and FrzZ and, as a consequence, into a high frequency of polarity switches and directional changes.
The M. xanthus community secretes a complex extracellular matrix that organizes multicellular movements and facilitates swarm expansion. At least two major exopolysaccharides (EPS and BPS) are secreted and both are essential for group movements, promoting Type-IV pilus motility and exerting a surfactant action to counteract surface tension. We study the function of this complex matrix as well as its structure during the predation cycle. The extracellular matrix is also coupled to motility by signaling networks in the form of a positive feedback loop that both enhances Type-IV pilus activity and EPS synthesis. We are studying how these systems interact at the molecular level.
Prey killing was long thought to involve the extracellular secretion of lytic enzymes and molecules by the predator. In fact, we recently discovered that killing depends mostly on predator-prey contacts involving a newly discovered pilus-like system (Tad) that becomes assembled specifically when Myxococcus cells establish contact with prey cells. We are currently studying how this system promotes killing in coordination with motility.
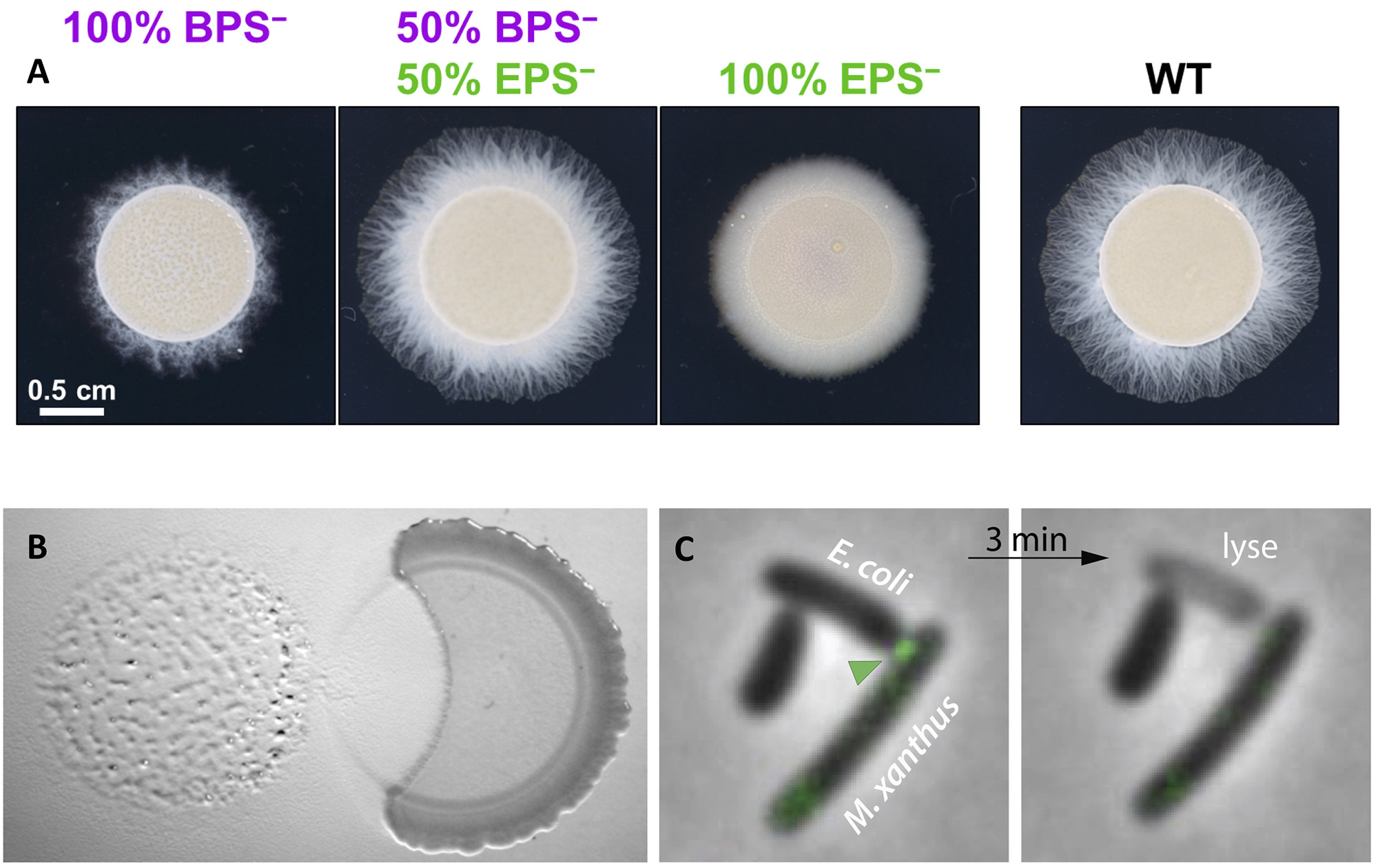
Figure legend : (A) Cross-complementation of EPS vs. BPS deficiencies via strain mixing. (A) EPS− and BPS− cells were mixed at a 1:1 ratio. Pure and mixed cultures were then spotted on soft agar and imaged after 48 h at 32 ºC. (B) Invasion and contact-dependent predation of a Bacillus colony (right) by Myxococcus (left). (C) Contact-dependent of E. coli by M. xanthus. Kil proteins (green arrohead) are recruted at the contact site between Myxococcus and its prey, immediately followed by the prey plasmolysis.
How does predation evolve in the soil? And what exact interactions does Myxococcus experience in its true ecological niche? We are now expanding our laboratory studies to field work and test how the mechanisms that we uncover in controlled laboratory conditions are used in the soils. For that, we are adopting a series of distinct approaches combining experimental evolution in the laboratory, omics approaches wild strains and, more recently, environmental sampling of Myxococcus interactions by metagenomics analyses of soil samples. Doing so, we hope to determine the ecological function of these bacteria and how they balance the composition of soil ecosystems. These studies could have direct applications for the restoration of biodiversity-deprived soils and understanding of plant-associated microbiota in the rhizosphere.
Laboratory director / Group leader / Research director (DR- CNRS)
Technician (TCS-CNRS)
Assistant professor (MCF-AMU)
Researcher (CR-CNRS)
Research engineer (IR-AMU)
Assistant engineer (AI-CNRS)
Researcher (CDD-CNRS)
Engineer (CDD-CNRS)
Researcher (CDD-CNRS)
Researcher (CDD-CNRS)
Researcher (CDD-CNRS)
Engineer (CDD-CNRS)
PhD student (PhD-AMU)
PhD student (PhD-AMU)
PhD student (PhD-AMU)
PhD student (PhD-CNRS)



