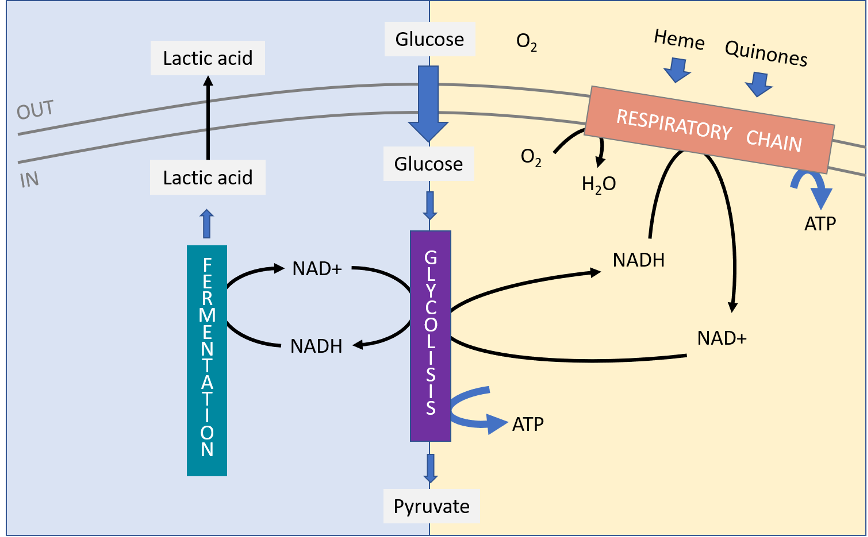
Science – Mise en évidence d’un nouveau module structural pour l’acheminement des quinones
Mise en évidence d’un nouveau module structural pour l’acheminement des quinones Pour fonctionner, les cellules doivent relier efficacement différents circuits métaboliques à travers des réactions