Job Opportunity – Postdoctoral Position in Microfluidics and Fluorescence Microscopy Applied to Cyanobacteria
Offre d’emploi – Ingénieur de recherche Microscopie & Microfluidique appliquées aux cyanobactéries Powered By EmbedPress
Our group studies cellular differentiation and pattern formation in cyanobacteria using the filamentous multicellular cyanobacterium Anabaena PCC 7120 as a model. When combined nitrogen becomes limiting, nitrogen-fixing cells called heterocysts differentiate from vegetative cells at semi-regular intervals along the filaments (every 10 to 15 vegetative cells). This one-dimensional pattern emerges following Turing-model rules. We have elucidated a key aspect of patterning of cyanobacterial heterocysts by uncovering the immunity mechanism that protects the developing cell from self-inhibition. Using genetics, cellular biology, and genomics, we aim to address major developmental questions of differentiation in Anabaena : How are the inhibitory morphogens processed and transferred along the filaments? What are the signaling networks that maintain the pattern? How does the circadian clock impact the development? What is the evolutionary history of the developmental genes across the cyanobacterial phylum?
We are also interested in engineering Anabaena in order to conciliate the oxygen-sensitive hydrogen production with photosynthesis. Since, Sunlight, CO2 and water are the only prerequisite to cyanobacteria growth, these organisms are candidate of choice for the production of biofuel.

Offre d’emploi – Ingénieur de recherche Microscopie & Microfluidique appliquées aux cyanobactéries Powered By EmbedPress
Bacterial RNA polymerase holoenzyme requires sigma70 factors to start transcription by identifying promoter elements. Cyanobacteria possess multiple sigma70 factors to adapt to a wide variety
Are you looking for an exciting and supportive team for your Master’s level internship? Are you passionate about environmental bacteria? If so, we have an
Huge congratulations to Dr Raphaël Rachedi who brilliantly defended his thesis on March 16th Thesis entitled : New insights into the regulation of heterocyst formation
Under nitrogen-limiting conditions, the filamentous cyanobacterium Nostoc PCC7120 differentiates nitrogen-fixing heterocysts at semi-regular intervals along filaments generating a periodic pattern of two distinct cell types.
Under combined nitrogen starvation, Anabaena perceives the presence of N2 and launches in response a developmental program leading to heterocysts: microoxic cells devoted to atmospheric nitrogen fixation. Heterocysts are formed at a semi-regular pattern that follows the Turing-model rules: in the developing cell, a transcriptional regulator (HetR) activates a specific transcriptional program that includes the expression of its own inhibitors. The diffusion of these inhibitory morphogens along the filament ensures pattern establishment. This initiation step is followed by a morphological differentiation which leads to the mature heterocyst and only at this point the nitrogenase encoding genes are expressed and N2 is fixed. We study the molecular mechanisms that allow the developing cell to sense the signal and to express the genes specific to each step of the process. How the transcriptional regulators involved act to orchestrate this temporal and cell specific transcription program? What are the sigma factors involved and how is their action regulated? How are the morphogens maturated and how do they diffuse along the filament? What is the distribution and evolution of the heterocyst-specific genes across the cyanobacterial phylum? Those are the main questions we aim to answer by combining genetics, biochemistry and genomics approaches.
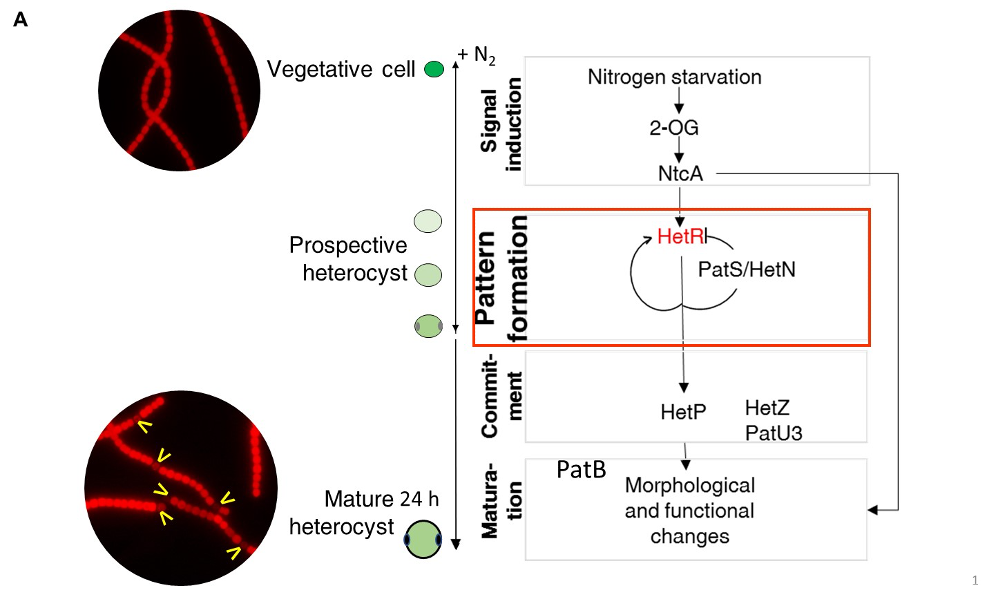
Legend FigA : The three main steps of cell differentiation in Anabaena: NtcA, HetR and PatB are transcriptional regulator controlling the heterocyst-specific transcriptional program. HetP, HetZ and PatU3 are involved int the commitment step. The activity of HetR in the vegetative cells is inhibited by diffusible morphogens issued from PatS, PatX and HetN. The yellow arrows indicate heterocysts.
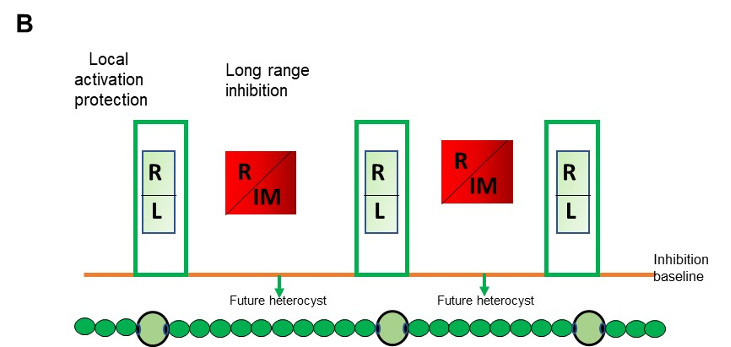
Legend FigB : Self-organized patterning in Anabaena: Local activation/protection: once activated, HetR activates the transcription of hetR, hetL, patS/hetN. The HetR/HetL network is favored. HetL provides immunity against the inhibiting morphogens produced in situ or entering the cell. HetR is active, heterocyst develops. Long range inhibition: the diffusion of the morphogens from both sides of the heterocyst creates an inhibition gradient. In the HetR/Morphogen network, HetR is inactive and the concentration of HetL is below the protective threshold. Differentiation is inhibited R: HetR, L: HetL or HetL homolog, IM: inhibiting morphogen (Providing from the processing of PatX, PatS or HetN).
Circadian rhythms are essential processes with broad implication on the physiology and adaptation of almost all living cell. Endogenous circadian clocks are critical to control temporal programs of cellular physiology to anticipate and adapt daily environmental changes like intensity light or temperature created by earth rotation. Although the molecular core clock functioning in a cell begins to be well understood in several organism, several key questions remain to be addressed. How do the circadian clock present in every cell of an organism are synchronized? How the same circadian clock function in different cell type with different transcriptional program? Robust and precise intracellular circadian timing has been described in cyanobacteria with positive impact on their fitness in rhythmic environment, and more particularly in a unicellular cyanobacterium. Our aim is to extend the study of the regulation of circadian rhythms to a multicellular bacterium, the cyanobacterium Anabaena. This filamentous bacterium constitutes a unique model to investigate, at the single cell level, the integration of the circadian rhythm into the differentiation cycle of a multicellular organism. By combining genetics, cell biology and modern age genome wide approaches, we are analyzing the molecular actors involved in timekeeping with an emphasis on the functioning of the clock in the heterocyst cell. Thus, addressing these questions in a relatively simple organism will help to understand the biological clock in a more complex tissue/organism.
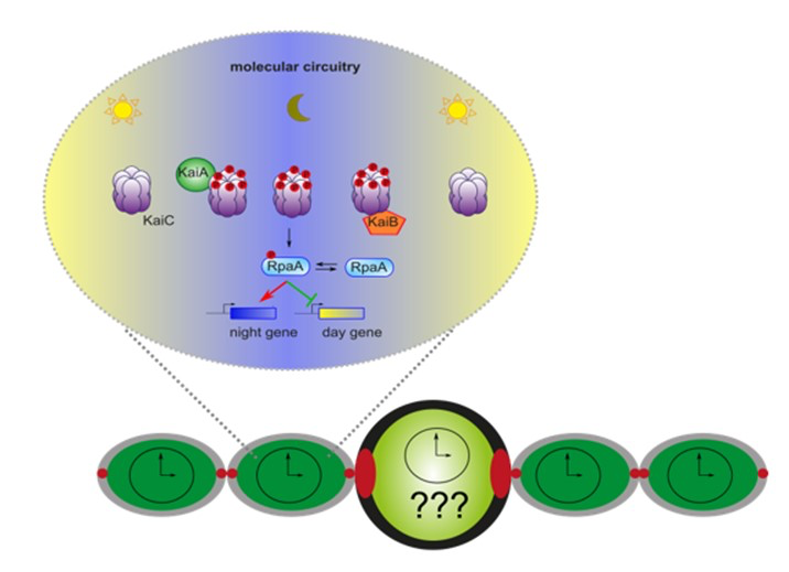
Legend Fig : A model for circadian clock functioning in cyanobacteria. KaiA, KaiB and KaiC constitute the core circadian clock. KaiA promotes the phosphorylation (red) of KaiC at dusk while KaiB has an antagonist activity. For the output, KaiC phosphorylates RpaA, a response regulator with the phosphoryl form (red) peaking at dusk. RpaA activates the expression of night genes and repress the expression of the day genes allowing rhythmic expression at a genome wide level. This model is based on experiment done in a unicellular cyanobacterium, Synechococcus elongatus. The goal of this axis is to show that the circadian clock is working similarly in the vegetative cell of Anabaena and to see if the clock is functioning similarly in a differentiated cell such as heterocyst cell.
Nitrogen fixation is the only biological entryway of new nitrogen into the open ocean and therefore constitutes a main driver of oceanic primary production. As a result, not only the present diazotrophic activity but also its possible changes in response to a future, warmer climate, as well as their impact on the ocean biogeochemistry are key parameters to determine for a correct estimation of the energy fluxes related to diazotrophic growth. To improve our knowledge of the actual representation of diazotrophs, we are scrutinizing the growth efficiency of the marine cyanobacterium Crocosphaera watsonii in regard to phosphorus and temperature changes by running tightly controlled and closely monitored culture experiments. To accurately evaluate the responses, at a molecular level, to rapid and transient changes in growth parameters, the transcriptional response of key genes involved in nitrogen fixation and response to nutrient starvation is evaluated.
Since nitrogen fixation produces hydrogen as a byproduct, diazotrophic cyanobacteria are considered as interesting candidates for Bio-H2 production. In addition, several strains produce hydrogenases that can also be used for H2-production. However, Oxygen sensitivity of nitrogenases and hydrogenases is a significant limit to the use of cyanobacteria as cell factories for H2-prodcution. We have shown that the heterocysts of Anabaena can be used a production-unit for O2-senstitive H2ase (HydA from Clostridium). More recently we have identified the presence of O2-tolerant H2ases in the genomes of some cyanobacteria. We are currently studying this family of enzyme in the unicellular cyanobacterium Cyanothece by answering the following questions: What is the physiological function of this enzyme? Is it connected to nitrogen fixation? What is the molecular basis of its O2-tolerance and would be used for high yield O2-production?
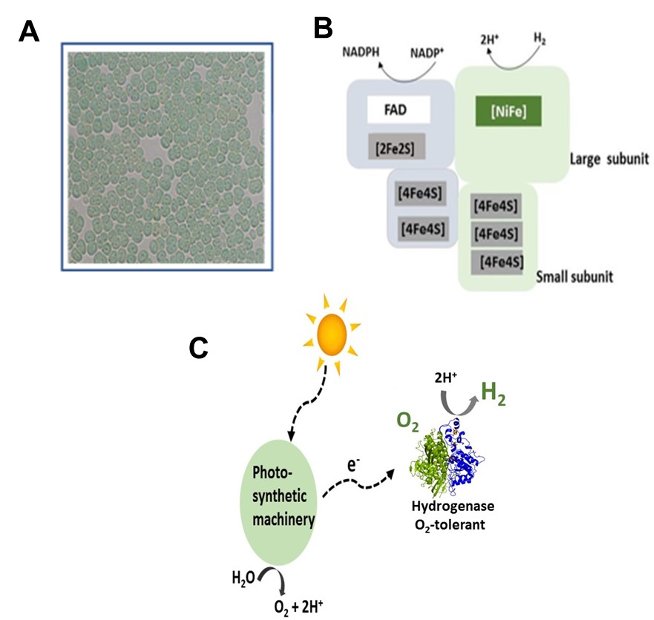
Legend Fig : Integration of hydrogen metabolism in a diazotrophic unicellular cyanobacterium A: Cyanothece PCC 7425 microscopic images. B: Schematic representation of the O2-tolerant hydrogenase. C: The potential O2-tolerance of the hydrogenase reconciles hydrogen production and phototrophic growth conditions.
Group Leader / Professor (PR-AMU)
Assistant professor (MCF-AMU)
Assistant professor (MCF-AMU)
Researcher (CR-CNRS)
Engineer (IE-AMU)
Engineer (CDD-CNRS)
PhD student (PhD-AMU)
PhD student (PhD-CNRS)
PhD student (PhD-AMU)



