HieVi: protein Large Language Model for proteome-based phage clustering
From S. Panigrahi, M. Ansaldi & N. Ginet
The “Phage Cycle and Bacterial Metabolism” team at Laboratoire de Chimie Bactérienne (Centre National pour la Recherche Scientifique/Aix-Marseille Université) is pleased to introduce its Phage Atlas :
https://github.com/pswapnesh/HieVi/raw/refs/heads/main/HieVi_UMAP.html
This Phage Atlas was generated with the Hierarchical Viruses (HieVi) pipeline we developed to explore the bacteriophage world diversity using machine learning-based representations. HieVi harnesses the Evolutionary Scale Model 2 (ESM-2) protein Language Model (pLM) to describe entire viral proteomes. We organized bacteriophages in a hierarchical tree that captures multiscale evolutionary relationships in good accordance with established bacteriophage phylogenies. HieVi is the first example of pLM implementation to sort and compare biological entities, opening new perspectives for comparative genomics for biologists in the field.
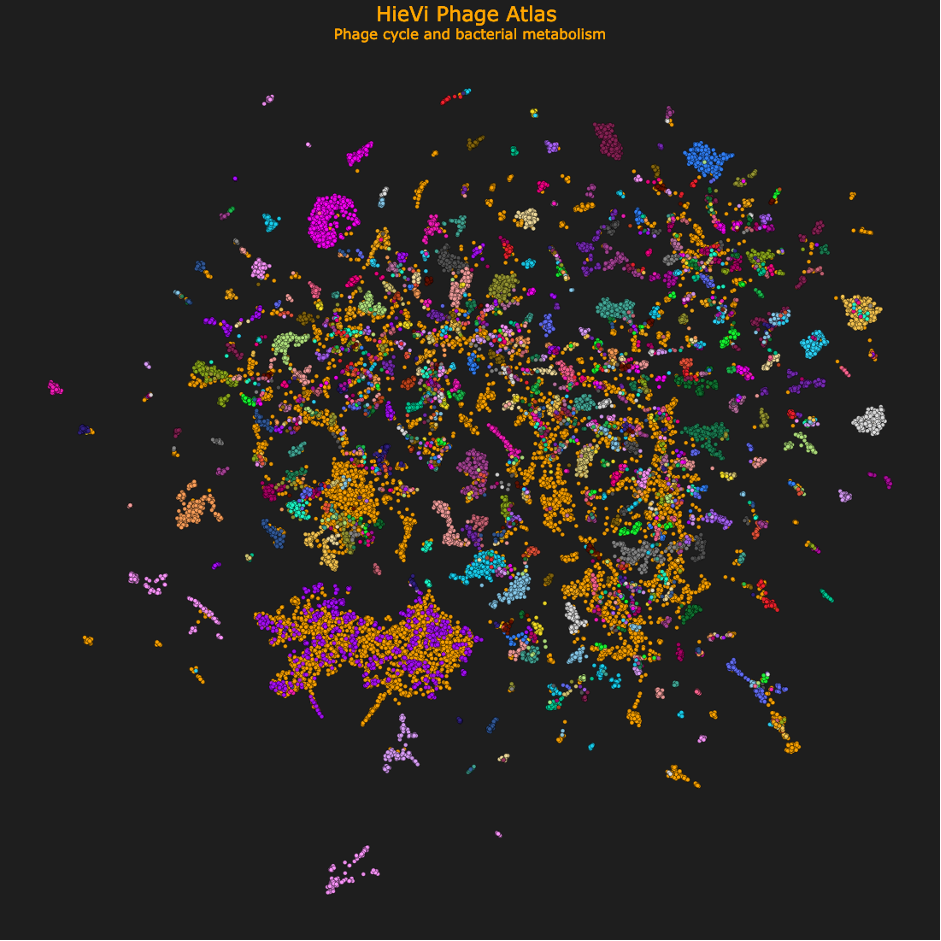
HieVi phage Atlas encompassing 24,362 complete and annotated viral genomes.



