Etienne Maisonneuve
Group leader / Researcher (CR-CNRS)
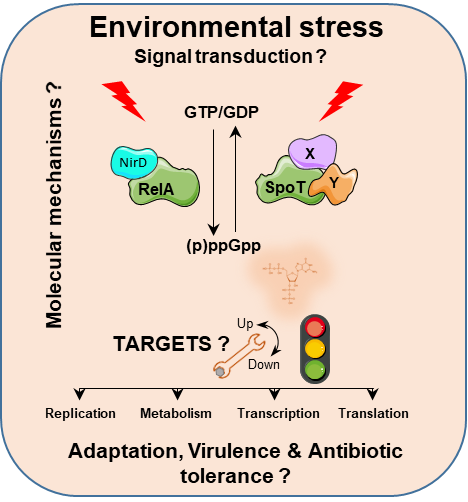
Bacteria have evolved various molecular mechanisms to cope with environmental stress, including the use of nucleotide-based signaling pathways to efficiently coordinate cellular processes and to provide a fast response. Among these signaling pathways the Stringent Response is a general bacterial stress response induced by diverse nutritional and environmental stresses that allows bacteria to rapidly survive adverse conditions. This global reprogramming of cells physiology relies on the accumulation of two unusual phosphorylated derivatives of GTP and GDP collectively called guanosine penta and tetra phosphate [(p)ppGpp] which are synthetized by the RelA/SpoT homologue (RSH) proteins. The molecular mechanisms by which environmental cues activate the stringent response are still largely unknown and represent an unsolved problem in prokaryotic molecular biology. Moreover (p)ppGpp has emerged as an important regulator of not only the bacterial stress response but also of many aspects of bacterial physiology including virulence, immune evasion, and antibiotic tolerance, strongly arguing that novel insights into (p)ppGpp biology will lead to novel methods to combat infections. By combining genetic and biochemical approaches we are deciphering the molecular mechanisms that trigger (p)ppGpp accumulation in the bacterial model Escherichia coli with special emphasis on their role in antibiotic tolerance.
The Stringent Response is a general bacterial stress response induced by diverse nutritional and environmental stresses allowing bacteria to adapt and survive adverse conditions. This global resetting of bacterial cells physiology relies on the accumulation of two unusual phosphorylated derivatives of GTP and GDP collectively called (p)ppGpp which are synthetized by the RelA/SpoT homologue proteins. Importantly the Stringent Response has a key role in bacterial virulence and in the formation of antibiotic-tolerant cells. However, the molecular mechanisms by which environmental cues activate the stringent response are still largely unknown and represent one of the most fundamental, unsolved problems in prokaryotic molecular biology. Thus, there is a pressing need to understand how the activities of the enzymes responsible for synthesis and degradation of (p)ppGpp are regulated and to develop novel strategies to combat recalcitrant infections. Therefore, the STRINGENCY project aims to uncover the molecular mechanisms by which environmental stimuli trigger SpoT-dependent (p)ppGpp synthesis with special emphasis on its role in the formation of multidrug tolerant cells in Escherichia coli. The STRINGENCY project was organized around three main objectives: (i) To unravel how spoT expression is regulated (ii) To reveal how (p)ppGpp synthetase II activity encoded by spoT is mechanistically controlled and (iii) To decipher the physiological role of SpoT in multidrug tolerance.
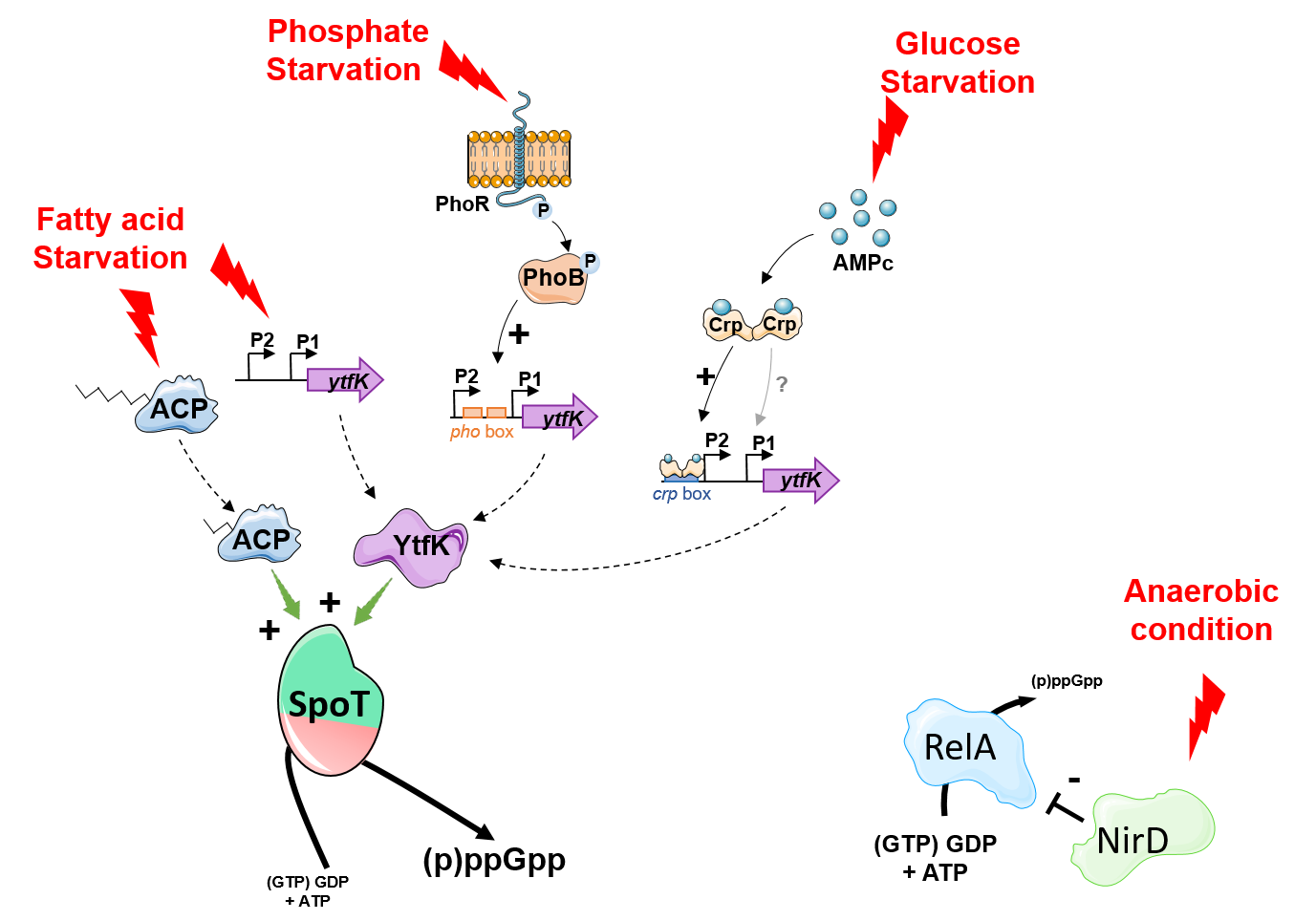
To gain further insights into the regulation of the bifunctional (p)ppGpp synthetase/hydrolase SpoT enzyme we have successfully developed genetic assays that allowed us to identify new protein candidates that modulate (p)ppGpp level in E. coli. We have isolated six candidates that either promotes accumulation or degradation of (p)ppGpp in vivo. Among them we observed that YtfK, a small protein of unknown function, is required to sustain elevated (p)ppGpp levels in response to phosphate, carbon and fatty acid starvation, therefore ensuring cell survival under these stress conditions. Interestingly YtfK is also needed to produce antibiotic tolerant cells under nutritional stress conditions, further highlighting the important role of SpoT and the signaling molecule (p)ppGpp in controlling multidrug tolerance. Mechanistically we show that YtfK controls the stringent response via a direct and specific interaction with the N-terminal region of SpoT encompassing the hydrolase and synthetase domains. We further observed that YtfK modifies the protein surface of SpoT catalytic domains supporting conformational changes upon binding and suggesting that YtfK acts by tilting the catalytic balance of SpoT toward (p)ppGpp synthesis rather than hydrolysis. In addition we decipher the genetic controls involved in the regulation of ytfK expression and its impact on the cell physiology. We observed that ytfK is subjected to catabolite repression and is positively regulated by the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex. Moreover we observed that YtfK controls SpoT-dependent accumulation of (p)ppGpp, cell survival and formation of antibiotic tolerant cells in response to carbon starvation. Finally we uncovered an unprecedented mode of regulation that inhibits the stringent response in Escherichia coli. Indeed, we discovered that the small subunit of the nitrite reductase NirD prevents (p)ppGpp accumulation in vivo from the mono-functional (p)ppGpp synthetase RelA. Remarkably, NirD modulates the stringent response by direct binding to RelA in vivo. Mechanistically, our in vitro results show that binding of NirD to the catalytic domains of RelA directly inhibits the rate of pppGpp synthesis.
Taken together, the identification of new partners that modulate the stringent response and the characterization of the molecular mechanisms behind these regulations have critically increase our knowledge of (p)ppGpp biology and bacterial survival mechanisms and may, in the future, improve biotechnological processes and how bacterial infections are treated.
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program grant agreement n° 71493 (STRINGENCY).

The stringent response is a general stress response that is mediated by the accumulation of the alarmone (p)ppGpp. These signaling molecules allow bacteria to rapidly respond and adapt to various conditions of nutritional and environmental stress by affecting gene expression and metabolism. In Escherichia coli, accumulation of theses alarmones depends on the (p)ppGpp synthetase RelA and the bifunctional (p)ppGpp synthetase/hydrolase SpoT. A tight regulation of these intracellular activities is therefore crucial to rapidly adjust the (p)ppGpp levels in response to environmental stresses. By using genetic screening approaches we have identified new proteins candidates that modulate the stringent response in E. coli (Germain et al., 2019) (Léger et al 2021. By combining genetic and biochemical approaches we are now deciphering the molecular mechanisms by which these candidates control (p)ppGpp homeostasis with special emphasis on their role in bacterial fitness and cell survival under fluctuating stress conditions.
The outcome of this basic research program has the potential to increase our understanding of (p)ppGpp biology and bacterial survival mechanisms and may, in the future, improve biotechnological processes and how bacterial infections are treated.
Group leader / Researcher (CR-CNRS)
Assistant professor (MCF-AMU)
Engineer (IE-CNRS)
PhD student (PhD-CNRS)
PhD student (PhD-AMU)



